In this guide, we will learn how to add measured data to a Salvus project. We
will then run some simulations by building a digital twin of the real-world
experiment and compare the actually measured data with simulated data. The
example that we will look at here is concerned with ultrasonic inspection. We
will use a reference ultrasonic dataset recorded on a polyamide block
specimen with a simple geometry. The simplicity of the experiment highly
facilitates the interpretation of the data. The dataset is described in
detail in the paper Maack et al. (2022), Low frequency ultrasonic dataset
for pulse echo object detection in an isotropic homogeneous medium as
reference for heterogeneous materials in civil
engineering.
The full dataset is published open-access under the Creative Commons license
and can be downloaded
here.
To keep the computational costs tractable, we will look at a select few shots
out of the dataset.
By the end of this tutorial, you will:
- Know how you can add your own measurement data to a Salvus project.
- Know how to use the Salvus internal waveform retrieval functionality to plot comparisons between observed and simulated data.
- Have learned how you can simulate data of (phased-)array transducer ultrasonic probes.
- Have observed how Salvus simulations in a digital twin can match real-world experimental data 'wiggle-by-wiggle'.
The polyamide block specimen used in the experimental data looks like this
(Figure reproduced after Figure 5 in Maack et al.
(2022)).
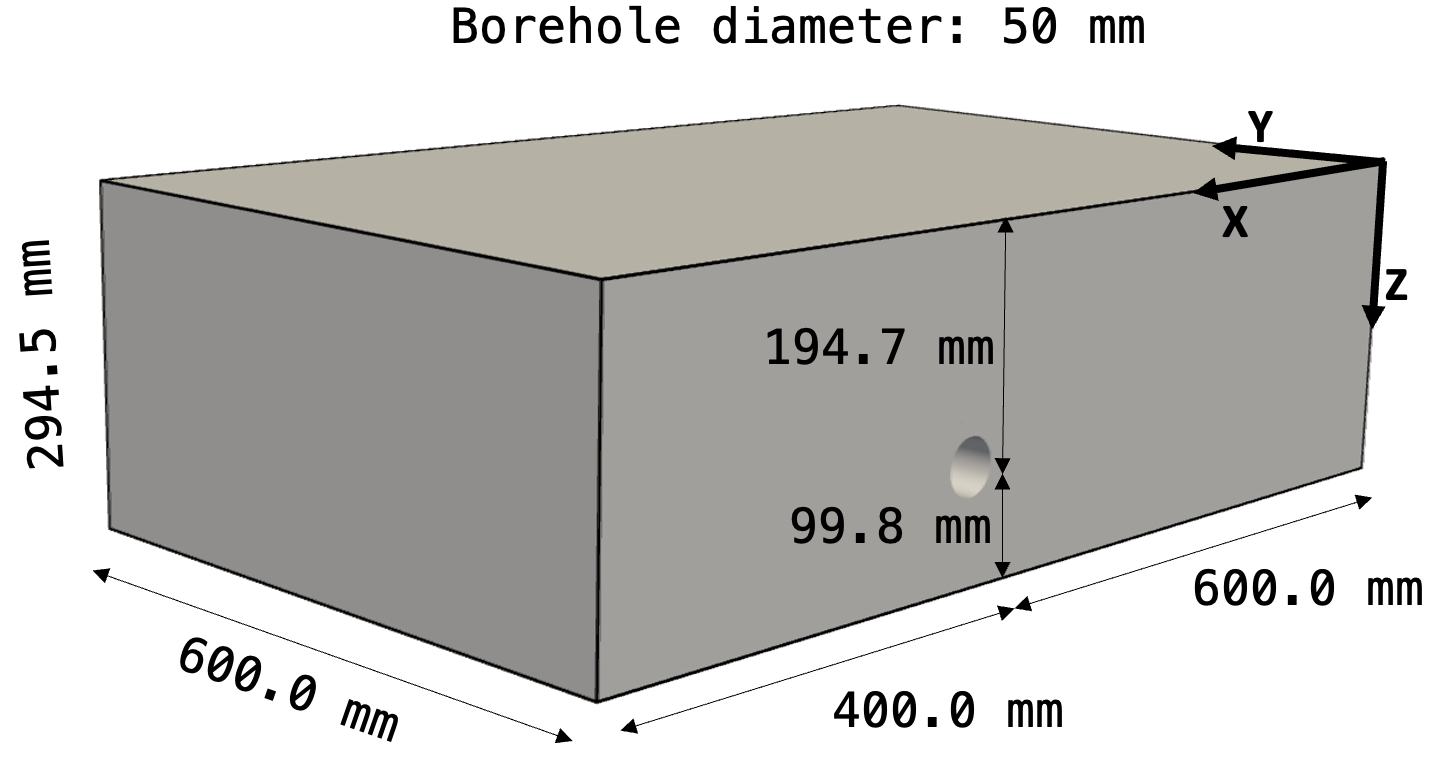
As you can see, the test block contains a single borehole with a diameter of
50 mm. The experimental data was recorded in the coordinate frame depicted by
the bold arrows with the origin in the top right corner. We now want to use
Salvus to simulate ultrasonic data in a digital twin of this test block and
compare it to the actually recorded data.
As always, we first import some of the Python packages that we need to run
this example.
import os
import matplotlib.pyplot as plt
import numpy as np
import xarray as xr
import salvus
import salvus.namespace as sn
SALVUS_FLOW_SITE_NAME = os.environ.get("SALVUS_FLOW_SITE_NAME", "local")Let us start by setting up a Salvus project from a simulation domain where we
set up the dimension object from the measures of the test body given in the
figure above. Note that, following Salvus coordinate conventions, the x- and
z-axes are flipped compared to the coordinate frame that the measurements are
defined in.
domain = sn.domain.dim3.BoxDomain(
x0=0.0,
x1=1.0,
y0=0.0,
y1=0.6,
z0=-0.2945,
z1=0.0,
)
p = sn.Project.from_domain(
path="project",
domain=domain,
load_if_exists=True,
)
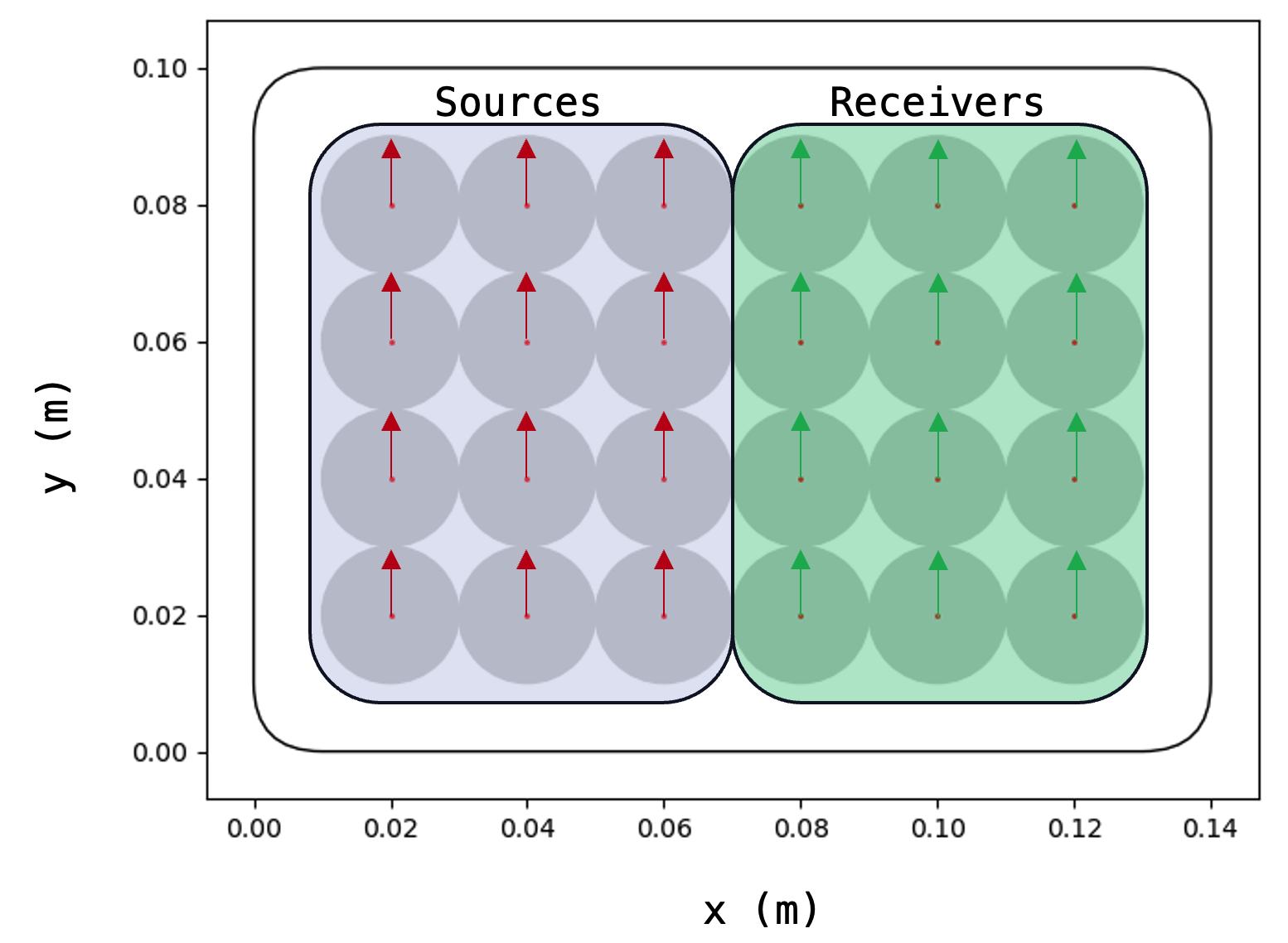
domain.plot()The data were recorded with an ultrasonic probe that includes 24 transducer
elements. From the 24 transducers, 12 are simultaneously excited as sources
and 12 are parallel-connected as receivers. This means that the signals
recorded by the 12 receivers are internally summed up to yield a single
output signal at each probe location.
Here we provide a view from the bottom of the probe where the small arrows
inside each transducer element indicate that the probe was run in Shear-wave
mode, meaning that transverse waves are excited in the direction of the arrow
at the source positions and the corresponding direction of particle
displacement is recorded at the receiver positions.
Now, let us load some of the measurement data. With this tutorial, we provide
the comma-separated file
'data/Shear_x001-x101_y025_Rot00.csv', which
contains a single line profile of data. Of course, please feel free to
download other profiles directly from the
source.
To acquire the line of data, the probe was moved across the surface of the
specimen in 10 mm increments in the x-direction. Such line measurements are
typically referred to as "B-scans". Each of the 100 columns of the dataset
thus corresponds to a single measurement position (indexed from 'x001 = 0 m' to 'x101 = 1 m'). The profile was located at a lateral position of
'y025=0.24 m'. For the B-scan included in this tutorial, the probe was
oriented in the following way (after Maack et al.,
2022):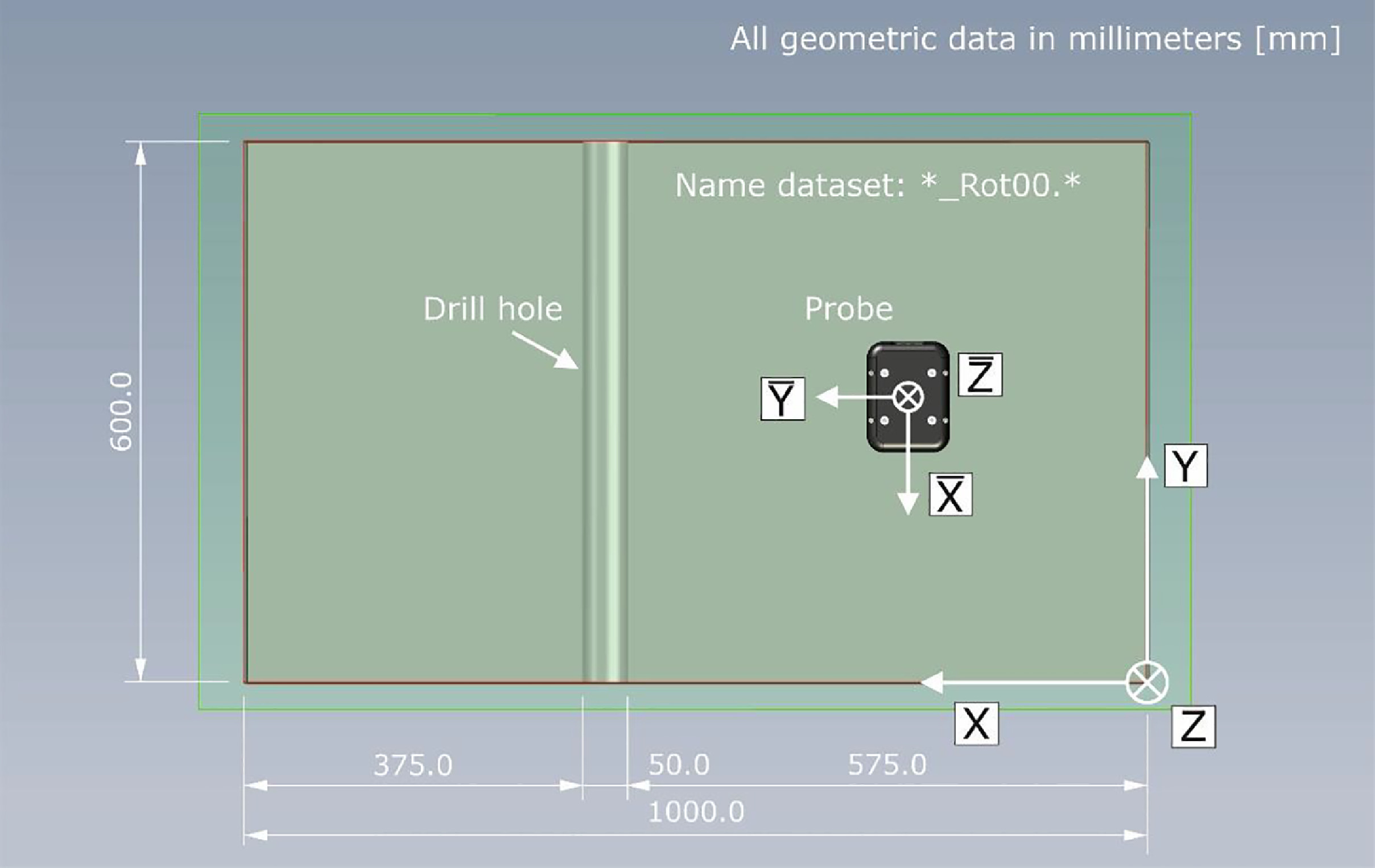
# Load the data from the CSV file
data = np.genfromtxt("data/Shear_x001-x101_y025_Rot00.csv", delimiter=",")
sampling_rate_in_hz = 2_000_000 # The data is sampled at 2 Mhz
dx = 0.01 # X-spacing of measurement positions
t = np.arange(data.shape[1]) * (1 / sampling_rate_in_hz) # The time axis
x = np.arange(data.shape[0]) * dx
y = 0.24 # Profile at y=0.24m
z = 0.0 # Measurements are taken at the surface at z=0mFor visualization, we now wrap this data into an
xarray.DataArray object
with an 'x' and 'time' coordinate, which provides us with a nice way to
store and plot structured data. We can already see some interesting features
in the data:- The direct wave close to t=0 seconds.
- The reflection off the bottom of the specimen at t~=5.5e-3 seconds and multiples thereof.
- The diffraction hyperbola of the borehole with the apex located at x=0.6 m.
observed_data = xr.DataArray(
data=data, dims=["x", "time"], coords={"x": x, "time": t}
)
observed_data.T.plot()<matplotlib.collections.QuadMesh at 0x722d449d82d0>
To perform data-fusion, we now would want to run Salvus simulations for the
same line of data. First, we define the material parameters of the polyamide
block. We have done some calibration of the material properties for you and
found the following properties to be a good representation of the physical
properties of the block:
- P-wave velocity: 1990 m/s
- S-wave velocity: 1136 m/s
- Density: 1150 kg/m^3
In practice, you could of course use Salvus' full waveform inversion
capabilities to calbirate the material properties, but this is
beyond the scope of this example.
The polyamide is also dissipative, so we will run a visco-elastic simulation
with the following attenuation parameters:
- Q factor for the bulk modulus: 85 m⁻¹
- Q factor for Lame's second parameter: 85 m⁻¹
Please refer to the attenuation
tutorial
for more information on running viscoacoustic and viscoelastic simulations
with Salvus.
From these properties, we set up a homogeneous model.
parameters = {
"VP": 1990.0, # P-wave velocity (m/s)
"VS": 1136.0, # S-wave velocity (m/s)
"RHO": 1150.0, # Density (kg/m^3)
"QKAPPA": 85.0, # Q for the bulk modulus.
"QMU": 85.0, # Q for Lame's second parameter (mu).
}
mc = sn.ModelConfiguration(
background_model=sn.model.background.homogeneous.IsotropicViscoElastic(
vp=parameters["VP"],
vs=parameters["VS"],
rho=parameters["RHO"],
qkappa=parameters["QKAPPA"],
qmu=parameters["QMU"],
),
linear_solids=sn.LinearSolids(reference_frequency=60_000),
)Now, we need to create Salvus
Event objects for each measurement position
on our profile, where an Event corresponds to a digital twin of the probe
transducer locations at a specific measurement position. So, let us first
specify some general geometrical properties of the probe.transducer_dx = 0.02 # Transducer spacing in x-direction
transducer_dy = 0.02 # Transducer spacing in y-direction
nx = 4 # Number of transducers in x direction for given probe orientation
ny = 6 # Number of transducers in y-direction for given probe orientation
transducer_x = np.arange(nx) * transducer_dx
transducer_y = np.arange(ny) * transducer_dy
X, Y = np.meshgrid(
transducer_x, transducer_y
) # XY Coordinates for each transducer
X, Y = X.ravel() - np.mean(transducer_x), Y.ravel() - np.mean(
transducer_y
) # Place the center of the instrument at the origin
# Split into sources and receivers
x_sources = X[Y > 0]
y_sources = Y[Y > 0]
x_receivers = X[Y < 0]
y_receivers = Y[Y < 0]
# A quick plot of the transducer coordinates for QC
plt.figure()
plt.plot(x_sources, y_sources, "b.", label="sources")
plt.plot(x_receivers, y_receivers, "g.", label="receivers")
plt.gca().set_aspect("equal")
plt.xlabel("X (m)")
plt.ylabel("Y (m)")
plt.title("Transducer locations")
plt.legend()<matplotlib.legend.Legend at 0x722d410b4bd0>
Now we define the Events for each measurement position by moving the
coordinates our device to the position of the measurements and defining the
corresponding sources and receivers. We do offer a digital twin library for a
series of commercially available ultrasonic probes that facilitates the
creation of these events. However, in this tutorial, we show how this can be
done manually in order to facilitate adapting your workflow to arbitrary
probe geometries.
for i, pos_x in enumerate(x):
event = sn.Event(
event_name=f"event{i+1:03d}",
sources=[
sn.simple_config.source.cartesian.VectorPoint3D(
x=1.0 - (sx + pos_x), # Account for the flipped x-axis
y=sy + y,
z=z,
fx=-1.0, # Horizontally oriented source, negative X-direction
fy=0.0,
fz=0.0,
)
for (sx, sy) in zip(x_sources, y_sources)
],
receivers=[
sn.simple_config.receiver.cartesian.Point3D(
x=1.0 - (rx + pos_x),
y=ry + y,
z=z,
network_code="XX",
station_code=f"AA{n:03d}",
fields=["displacement"],
)
for n, (rx, ry) in enumerate(zip(x_receivers, y_receivers))
],
# The concept of ReceiverChannel objects is described in the cell below
receiver_channels=[
sn.ReceiverChannel(
identifier="OUT",
input_receivers=[
f"XX.AA{n:03d}." for n in range(len(x_receivers))
],
input_receiver_weights=np.ones_like(x_receivers),
input_receiver_delays=None,
)
],
)
try:
p.add_to_project(event)
except ValueError:
# For some recording positions, the transducers would be located
# outside the domain. We do not consider these events here.
print(
f"event{i+1:03d} was not added to the project as some of the "
"sources and receivers lie outside the domain!"
)event001 was not added to the project as some of the sources and receivers lie outside the domain! event002 was not added to the project as some of the sources and receivers lie outside the domain! event003 was not added to the project as some of the sources and receivers lie outside the domain! event099 was not added to the project as some of the sources and receivers lie outside the domain! event100 was not added to the project as some of the sources and receivers lie outside the domain! event101 was not added to the project as some of the sources and receivers lie outside the domain!
Note that in addition to the source and receiver objects, we have set up a
ReceiverChannel object for each Event. Salvus ReceiverChannel's are a
convenient way to handle parallel-connected receivers, such as the ones of
our ultrasonic inspection probe. When we access the data later on, Salvus
will automatically form a weighted sum of the data of the 12 receivers to
combine them into a single output channel. A ReceiverChannel object takes
the following input arguments:identifier: A unique string that identifies the channel. Here, we simply used the identifier"OUT".input_receivers: A list of the receiver names that will be combined into a single output channel.input_receiver_weights: An array of the same length asinput_receiversthat specifies the relative weight that each receivers gets in the weighted sum output. In this example, we give each receiver the same weight.input_receiver_delays: An optional array of the same length asinput_receiversthat specifies a time delay in seconds by which the signal of each receiver is shifted before forming the weighted sum. This can be useful when you want to simulate data of phased-array probes. In this example, we do not specify any time delays.
We are now almost ready to run some simulations. What is still missing is the
source time function of the probe (i.e., the pulse that the sources
transmit). Here, we provide a calibrated source time function that is close
to the actual pulse emitted by the probe. The source time function is stored
in
"/data/stf.h5". Let us load and inspect it.stf = sn.simple_config.stf.Custom(filename="data/stf.h5", dataset_name="stf")
stf.plot()We now have all the information that we need to set up our simulation! We
will simulate 0.001 seconds of data in a viscoelastic setup. The mesh is
resolved with 1.3 elements per wavelength at a maximum frequency of 60kHz.
ec = sn.EventConfiguration(
wavelet=stf, # The source time function
waveform_simulation_configuration=sn.WaveformSimulationConfiguration(
end_time_in_seconds=0.001, # Total simulation time,
attenuation=True, # We want to run a visco-elastic simulation
),
)
# Combine everything into a complete configuration.
p += sn.SimulationConfiguration(
name="simulation",
elements_per_wavelength=1.3,
max_frequency_in_hertz=60_000,
model_configuration=mc,
event_configuration=ec,
)Let us verify that we have set up everything properly. We only visualize two
events (measurement positions) here. One at the very beginning of the profile
and one right above the location of the borehole.
p.viz.nb.simulation_setup(
simulation_configuration="simulation",
events=["event015", "event061"],
)[2026-01-29 01:22:11,596] INFO: Creating mesh. Hang on.
<salvus.flow.simple_config.simulation.waveform.Waveform object at 0x722d41161390>
This looks alright, so let us launch the simulations for these two events!
The simulations are relatively expensive in 3D, so ideally, we would run them
on a GPU.
p.simulations.launch(
simulation_configuration="simulation",
events=["event015", "event061"],
site_name=SALVUS_FLOW_SITE_NAME,
ranks_per_job=4,
)
p.simulations.query(block=True)[2026-01-29 01:22:12,582] INFO: Submitting job array with 2 jobs ...
True
Once the simulation runs are completed, we can retrieve the simulated data
for these two events from our project. Here, we directly retrieve the data as
xarray.DataArray objects If you are not familiar with xarray.DataArray
objects, please refer to our structured data
tutorial.
Note that get_waveform_data_xarray() automatically detects that our event
contains a ReceiverChannel object and only returns the summed data for the
receiver channel "OUT" rather than returning the data of the 12 individual
receivers. We only select the "X" component of the data here as this is the
only component that is measured by our ultrasonic probe given the probe
orientation. For the necessity of the specific xarray's functions used, it is
strongly suggested to also follow the xarray.DataArray tutorial.edc = p.waveforms.get(
"SYNTHETIC_DATA: simulation | normalize", events=["event015", "event061"]
)
data_event015 = (
edc["event015"]
.get_waveform_data_xarray(receiver_field="displacement")
.unstack()
.sel(component="X")
)
data_event061 = (
edc["event061"]
.get_waveform_data_xarray(receiver_field="displacement")
.unstack()
.sel(component="X")
)
# Print some information on the xarray data structure for the data of the first
# Event
data_event015<xarray.DataArray 'displacement' (time: 2088, receiver_id: 1)> Size: 8kB
array([[ 0.0000000e+00],
[ 0.0000000e+00],
[-4.4336452e-22],
...,
[-6.1103110e-03],
[-6.3030026e-03],
[-6.2529165e-03]], dtype=float32)
Coordinates:
* receiver_id (receiver_id) <U3 12B 'OUT'
component <U1 4B 'X'
* time (time) float64 17kB -0.0002044 -0.0002039 ... 0.0009994 0.001
Attributes:
sampling_rate_in_hertz: 1732756.0120452112
start_time_in_seconds: -0.00020443962421268225
reference_time_in_seconds: 0.0<xarray.DataArray 'displacement' (time: 2088, receiver_id: 1)> Size: 8kB
array([[ 0.0000000e+00],
[ 0.0000000e+00],
[-4.4336452e-22],
...,
[-6.1103110e-03],
[-6.3030026e-03],
[-6.2529165e-03]], dtype=float32)
Coordinates:
* receiver_id (receiver_id) <U3 12B 'OUT'
component <U1 4B 'X'
* time (time) float64 17kB -0.0002044 -0.0002039 ... 0.0009994 0.001
Attributes:
sampling_rate_in_hertz: 1732756.0120452112
start_time_in_seconds: -0.00020443962421268225
reference_time_in_seconds: 0.0# Now plot the X-component data for the two events
data_event015.plot(label="Event015")
data_event061.plot(label="Event061")
plt.legend()<matplotlib.legend.Legend at 0x722ccca97110>
The data for the two events look quite similar, which is not surprising
given the homogeneous properties of the block, the geometry of the probe and
the measurement positions. The additional pulse (at about 0.0003 seconds)
that can be seen in the data of Event015 corresponds to the reflection from
the right sidewall at x=1.0 meters.
To compare the simulation results with the recorded data, it is convenient to
add the experimental measurements to our Salvus project as well. In this way,
we can easily plot the simulated and observed data in the same way and use
some of Salvus' data filtering and normalization functions to facilitate the
comparison. Adding external data is of course also a crucial step for imaging
with full waveform inversion.
The easiest way to add external data to a Salvus project is by using the
function
salvus.data.external_data_to_hdf5(). It takes external data as a
structured xarray object and converts it to the HDF5 format that is
required by Salvus. This function will only convert the data, not add it yet
to the project on disk.Let us first try that for the data of a single event (here,
"Event015"). To
pass the data on to salvus, we need to prepare the data as a 3D xarray
structure with a specific format. The structure needs to have the three
following three dimensions:"receiver_id""component""time"
At each measurement position, we only have a single output signal
corresponding to combined signals of the 12 receivers described by the
receiver channel
"OUT" for the component "X". We can thus set up our data
in the following way, where we additionally pass on the attributes "dim"
(The number of dimensions considered in our project) and set the name of the
DataArray to the "receiver_field" (in our case, "displacement"):# Here we select the 15th (indexed by 14 when counting with 0) column of our
# observed data, corresponding to "event015"
dobs_event015 = observed_data.isel(x=14)
da = xr.DataArray(
data=np.reshape(
dobs_event015.data, (1, 1, len(dobs_event015.data))
), # The data as a 3D numpy array
dims=["receiver_id", "component", "time"], # The required dimensions
coords={
"receiver_id": ["OUT"], # The receiver identifier
"component": ["X"], # The component our data corresponds to
"time": observed_data.time, # The time axis
},
attrs={"dim": 3},
name="displacement",
)We now want to add those data to our project. To do so, we can use the
function
p.waveforms.add_external(). It will first validate that all
required information is available and that the data are compatible with the
project. Finally, it will add the data to the project.p.waveforms.add_external(
data=da,
data_name="external_data",
event=p.events.get("event015"),
)Now that the data is safely stored within our project, we can retrieve it
again and plot it.
# Retrieve an EventData object
ed = p.waveforms.get(
"EXTERNAL_DATA:external_data | normalize", events="event015"
)[0]
# Retrieve the data as an xarray.DataArray and select the "X" component
da = (
ed.get_waveform_data_xarray(receiver_field="displacement")
.unstack()
.sel(component="X")
)
# Plot the data
da.plot()[<matplotlib.lines.Line2D at 0x722cd441a3d0>]
As you can see, the data seems to contain some low-frequency noise — we
will deal with this in a second. First, let us also add the experimental data
of all the remaining measurement positions on our profile to the project. For
this, we simply loop over the Events and add the data in the same way we did
before.
for event in p.events.list():
# get the event number from the event name (the last three letters of the
# string)
id = int(event[-3:])
# The data is defined with 1-base indexing while Salvus is using 0-base
# indexing.
dobs = observed_data.isel(x=id - 1)
da = xr.DataArray(
# The data as a 3D numpy array
data=np.reshape(dobs.data, (1, 1, len(dobs.data))),
# The required dimensions
dims=["receiver_id", "component", "time"],
coords={
"receiver_id": ["OUT"], # The receiver identifier
"component": ["X"], # The component our data corresponds to
"time": observed_data.time, # The time axis
},
attrs={"dim": 3},
name="displacement",
)
if not p.waveforms.has_data(
"EXTERNAL_DATA:external_data", event
): # Only add the data if it has not already been added
p.waveforms.add_external(
data=da,
data_name="external_data",
event=p.events.get(event),
)For comparison, we now want to filter both the simulated and the experimental
data to the same frequency range to get rid of the low-frequency noise in the
observed data. To do so, we first define a filtering function that we can
pass to Salvus that bandpass filters our data between 40kHz and 60kHz. We
use a filtering function that uses Obspy under the hood to filter the data.
The function needs to have a specific signature as described in our
reference
documentation.
def filter_data(st, receiver, sources):
return st.filter(
"bandpass", freqmin=40_000, freqmax=60_000, zerophase=True
)We can add this processing function to our project by defining a
ProcessingConfiguration.pc = salvus.project.configuration.processing.ProcessingConfiguration(
name="bandpass",
data_source_name="EXTERNAL_DATA:external_data",
processing_function=filter_data,
)
p += pcWe now retrieve the processed data, clearly showing the effect of the filter.
edc_obs = p.waveforms.get(
"PROCESSED_DATA:bandpass | normalize", events=["event015", "event061"]
)
plt.figure()
da_obs_event015 = (
edc_obs["event015"]
.get_waveform_data_xarray(receiver_field="displacement")
.unstack()
.sel(component="X")
)
da_obs_event061 = (
edc_obs["event061"]
.get_waveform_data_xarray(receiver_field="displacement")
.unstack()
.sel(component="X")
)
# Plot the data
da_obs_event015.plot(label="event015")
da_obs_event061.plot(label="event061")
plt.legend()
plt.title("Experimental data")Text(0.5, 1.0, 'Experimental data')
Let us now compare the experimental data with the simulated data. For this,
we retrieve the simulated data in a similar fashion.
# Retrieve an EventData object
edc_sim = p.waveforms.get(
"SYNTHETIC_DATA:simulation | normalize", events=["event015", "event061"]
)
# Retrieve the data as an xarray.DataArray and select the "X" component and
# flip the sign to account for the different coordinate convention of the
# observed data
da_sim_event015 = -1 * edc["event015"].get_waveform_data_xarray(
receiver_field="displacement"
).unstack().sel(component="X")
da_sim_event061 = -1 * edc["event061"].get_waveform_data_xarray(
receiver_field="displacement"
).unstack().sel(component="X")
# Plot the data
plt.figure()
da_sim_event015.plot(label="event015")
da_sim_event061.plot(label="event061")
plt.legend()
plt.title("Simulated data")Text(0.5, 1.0, 'Simulated data')
Now we plot everything together.
plt.figure(figsize=(15, 5))
plt.subplot(121)
da_obs_event015.plot(c="k", label="experimental")
da_sim_event015.plot(c="r", label="simulated")
plt.legend()
plt.gca().set_xlim(0.0, 0.001)
plt.title("Event015")
plt.subplot(122)
da_obs_event061.plot(c="k", label="experimental")
da_sim_event061.plot(c="r", label="simulated")
plt.legend()
plt.gca().set_xlim(0.0, 0.001)
plt.title("Event061")Text(0.5, 1.0, 'Event061')
Note that the simulations seem to fit the data remarkably well! However,
there appear to be some minor differences between the simulated and the
observed data. Can you guess where these differences come from? The
explanation is simple. We did not include the borehole in our simulations. We
simply assumed the whole block is homogeneous. The difference is therefore
particularly visible for
"event061" that corresponds to a measurement
position directly above the borehole. We spare you from now running the
simulations again with the borehole as this would require a more
sophisticated meshing workflow — please refer to our meshing tutorials for
this. However, we already did the work for you and repeated the simulations
above for a mesh that includes the borehole. Check out the results in the
video below, showing both the volumetric wavefield and a comparison of the
experimental and simulated data. Note that we are now able to fully match the
data "wiggle-by-wiggle"!PAGE CONTENTS
 Mondaic Documentation
Mondaic Documentation